Analyses
The Alignment Resource
An Alignment is an analysis providing sensitive local alignment of a WGS samples against a specific reference genome sample. It has the same properties as an analyses, but lacks the 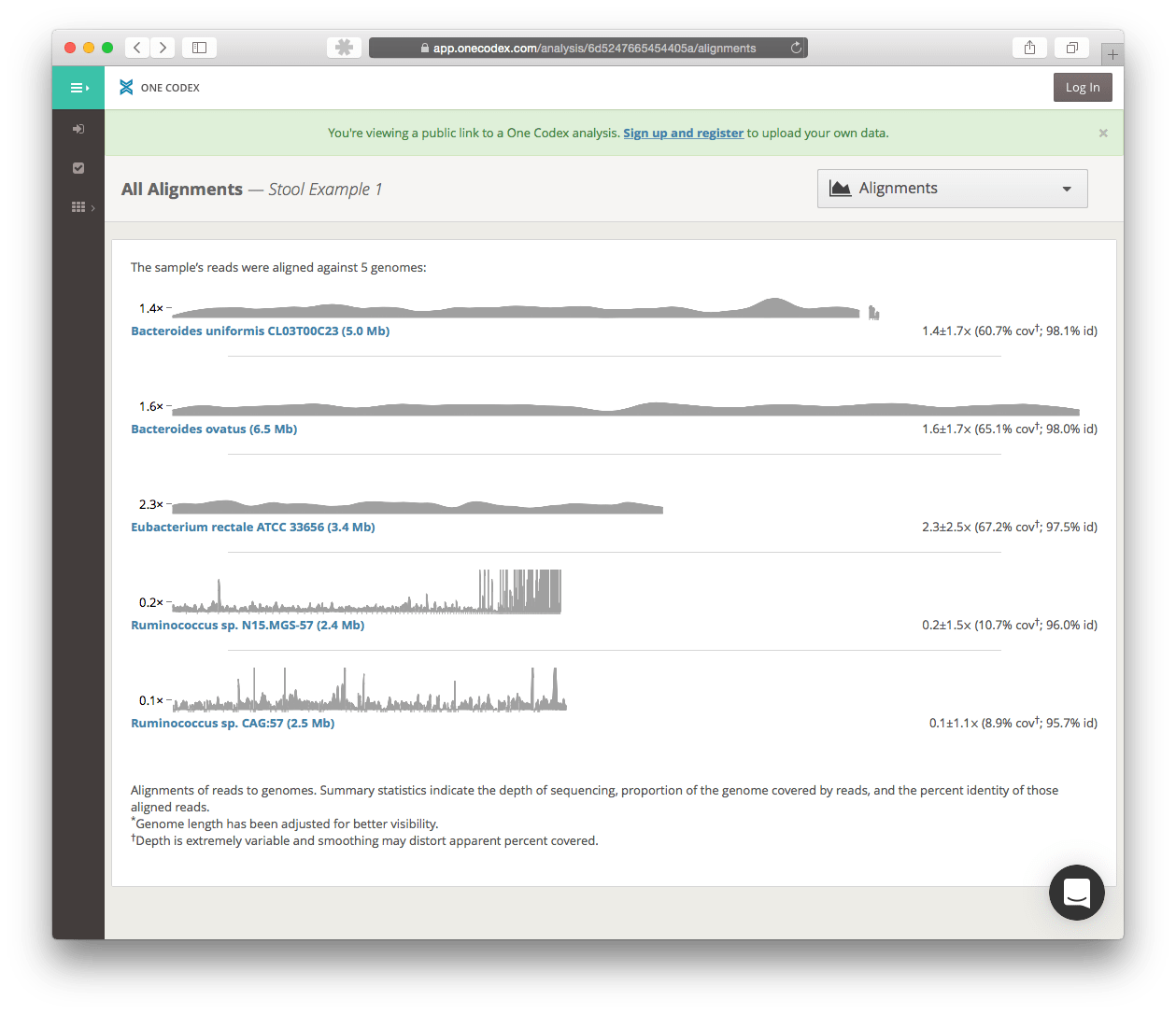
analysis_type field and also includes a tax_id field indicating the taxonmic ID of the genome against which the alignment was performed.
At this time, there are no v1 API routes for accessing the alignment statistics (e.g., percent identity, coverage, and depth), but alignments can be viewed on the One Codex platform directly. Here’s an example public alignment and a list of all alignments for that sample:

Pattern:
^/api/v1/alignments\/[^/]+$Example:
"/api/v1/alignments/a1b2c3d4e5f67890"
Maximum string length:
255A reference to the versioned job underlying the analysis, e.g., {"$ref": "/api/v1/jobs/d512cb556241440f"}.
The arguments passed into this analysis (can be null).
A reference to the sample underlying the analysis, e.g., {"$ref": "/api/v1/sample/0ee172af60e84f61"}.

