The Metadata Resource
"/api/v1/metadata/a1b2c3d4e5f67890"
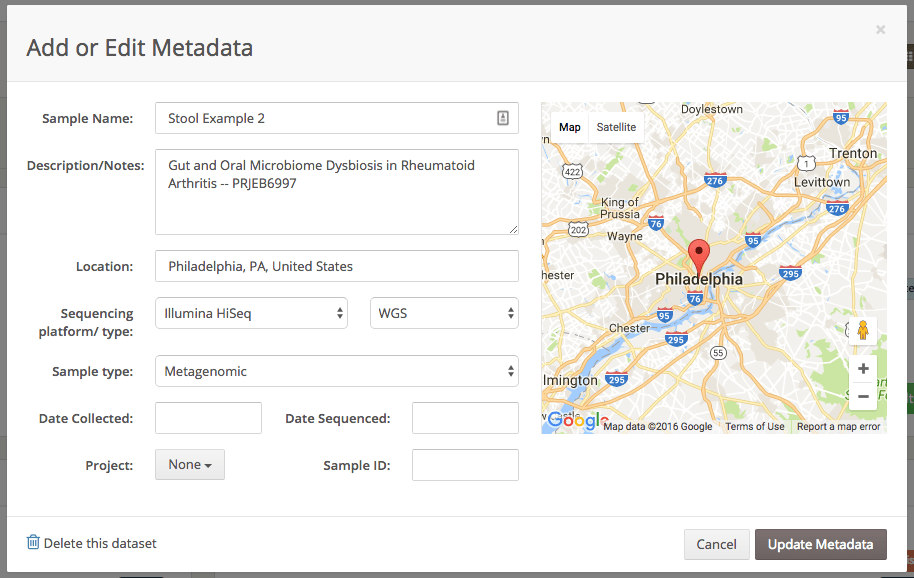
Arbitrary metadata is supported as part of a custom object. custom has two constraints: (1) it must have a depth of one (i.e., no nested records); and (2) only strings, numbers, boolean, and null values are supported as values. Example: {"lab_tech": "Linus Pauling", "amplicon_scheme": "V3-V4"}
Timestamp for when the sample was collected.
Timestamp for when the sample was sequenced.
An arbitrary external sample ID, e.g., an ID in a LIMS. Up to 60 characters.
60"a1b2c3d4e5f67890"
An enum with the sample library type.
WGS, Targeted/16S, Other, null The latitude (-90.0-90.0) of the sample location. By convention, we recommend using this for the location in which the physical specimen was collected.
-90 <= x <= 90The longitude (-180.0-180.0) of the sample location.
-180 <= x <= 180255255An enum with the name of the sequencing platform.
454 sequencing, 454 GS FLX, 454 GS FLX Titanium, Illumina, Illumina Genome Analyzer II, Illumina HiSeq, Illumina HiSeq 1500, Illumina HiSeq 2000, Illumina HiSeq 2500, Illumina HiSeq 3000, Illumina HiSeq 4000, Illumina HiSeq X, Illumina iSeq 100, Illumina MiniSeq, Illumina MiSeq, Illumina NextSeq, Illumina NextSeq 500, Illumina NextSeq 550, Illumina NovaSeq, Illumina NovaSeq 5000, Illumina NovaSeq 6000, BGISEQ, BGISEQ 50, Element, Element AVITI, Ion, Ion S5, Ion S5 XL, Ion PGM, Ion Proton, Ion Torrent, Oxford Nanopore, Oxford Nanopore GridION X5, Oxford Nanopore MinION, Oxford Nanopore PromethION, PacBio, PacBio RS II, PacBio Sequel, PacBio Sequel II System, PacBio Revio, PacBio Onso, SOLiD, Solexa Genome Analyzer, Sanger, Other, null The sample the metadata belongs to.
An enum with the sample type.
Isolate, Metagenomic, Other, null Whether the sample has been starred by the user within the One Codex web application.

