Skip to main contentWelcome to the One Codex API!
One Codex is a data platform for microbial genomics, designed for the secure storage, sharing, and reproducible analysis of microbial next-generation sequencing data. Our API powers our web application and is intended to be simple enough for use in ad hoc bioinformatic analyses, but powerful enough to support integration into existing systems (e.g., LIMS) or extension by building applications directly atop the platform.
More concretely, the API allows users to upload NGS data (FASTA or FASTQ files), automatically determine their composition them using our best-in-class metagenomic classifier and database, start new analyses such as in silico panels and alignments, retrieve and build on top of these analyses, and much more.
Core Concepts
Across the API, there are several core concepts which map directly to the endpoints detailed below:
-
Samples: A Sample is a collection of genetic sequences (reads or contigs) that can be uploaded for analysis on the One Codex platform. At this time, we support uploading both FASTA and FASTQ files, which can optionally be compressed using
gzip. Samples are owned by a User and may be described with Metadata records (both structured and free-form), annotated with Tags, and organized and shared via Projects.
-
Analyses: An analysis of an uploaded Sample. Several types of Analyses are supported on the platform today, and range from metagenomic classification (example) to panels for anti-microbial resistance (example). Types of analyses currently exposed via the API include Classifications, Panels, and Alignments. More are planned for the future.
-
Jobs: Analysis jobs represent an exact execution environment, analytic workflow, and accompanying reference data. Analyses are the result of a Job running against a specific Sample. All Jobs on the One Codex platform are strictly versioned and provide strong reproducibility guarantees, ensuring that an analyses of different samples can be readily compared and an analysis of the same sample can be repeated and reliably reproduced.
API Design & Accessing the API
Our API is a RESTful JSON API, and is self-described using JSON Schema. We hope that this format combined with an interactive API browser makes exploring and getting started with our API easy.
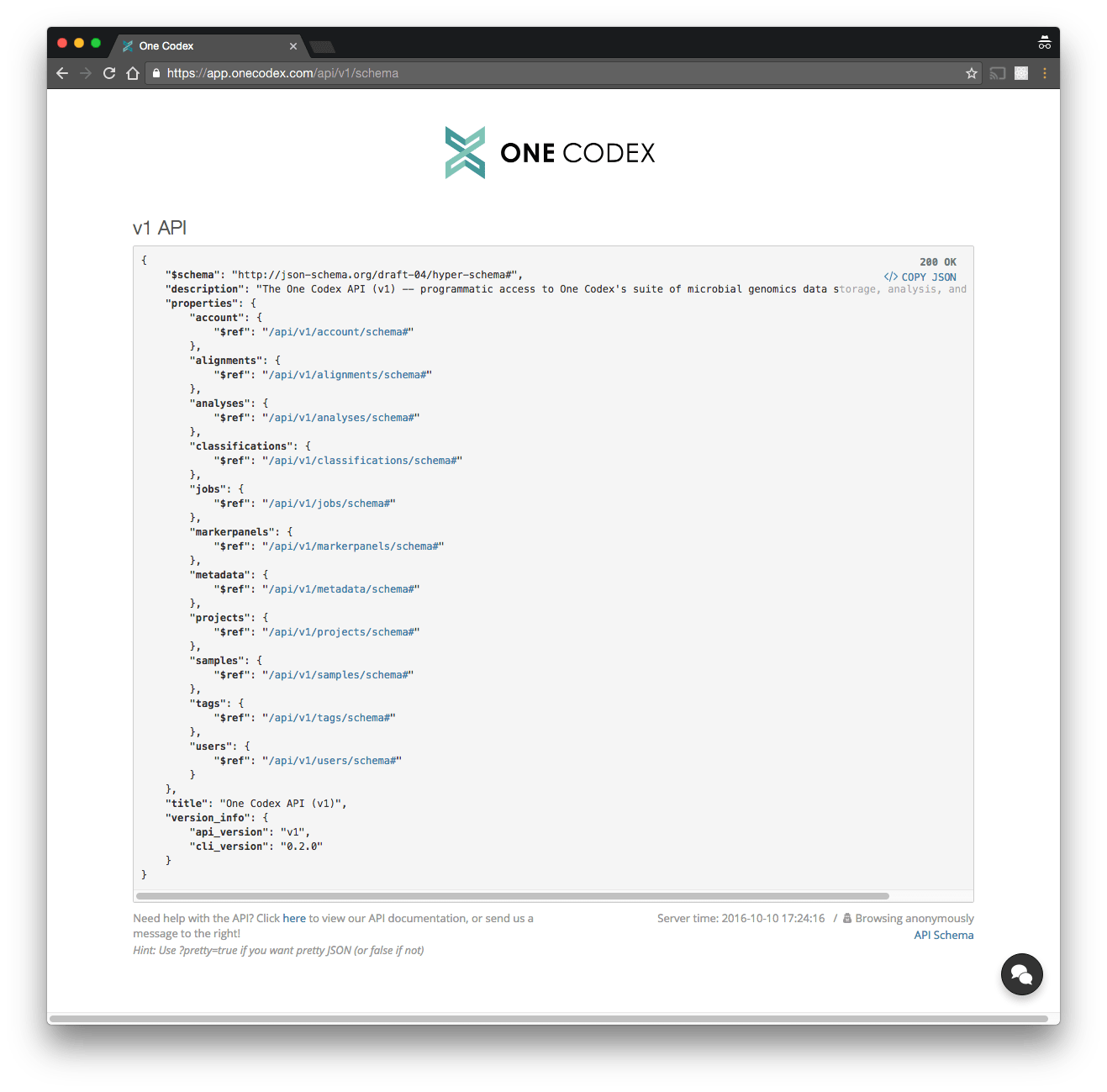 You can access the API using
You can access the API using cURL, an HTTP client library in your preferred language, our Python client library, or our command line client.
Finally, if you find any part of the docs to be outdated or unclear, please drop us a note and let us know! We’d also be grateful to hear any suggestions, requests, or other questions you have. You’re also always welcome to send us a note via the Chat button on the bottom right of these documentation pages and our API browser. 

